Wheat provides about one-fifth of the calories consumed by humans worldwide. About 95% of the wheat grown is common wheat, whereas most of the remaining are durum wheat. These wheats are traditionally characterized by a few quality properties and used for different end-use products, such as making bread or pasta, respectively. How were they linked in the domestication process and genomes have long been argued.
The puzzle of polyploid wheat evolution
The durum wheat is tetraploid (2n=4x=28, BBAA), which is thought be originated from Fertile Crescent more than 10,000 years ago. While the hexaploid (2n=6x=42, BBAADD) common wheat is thought to be originated as a result of polyploidization after the hybridization between domesticated tetraploid wheat (BBAA) and the goat grass (DD). The durum wheat and bread wheat had been cultivated together for a long time in history1, and the hexaploid common wheat took over durum wheat and is the most widely planted crop in the world.

The spreading history of wheat. Yield data: UN Food and Agriculture Organization
How did durum wheat and bread wheat connect in the gene pool? And how was the polyploid wheat successfully domesticated to be the most widely planted crop? These are important questions for understanding the evolution of domesticated wheat. Because interploidy cultivation of wheats is a traditional practice, hybridization occurs between wheat cultivars. However, the current understanding of wheat evolution could not fully explain the reticulate evolutionary process among wheats, which evolved with the pervasive hybrid swarm, gene flow and interploidy introgression.
Integrate statistic model and algorithm to decode the wheat complex genome
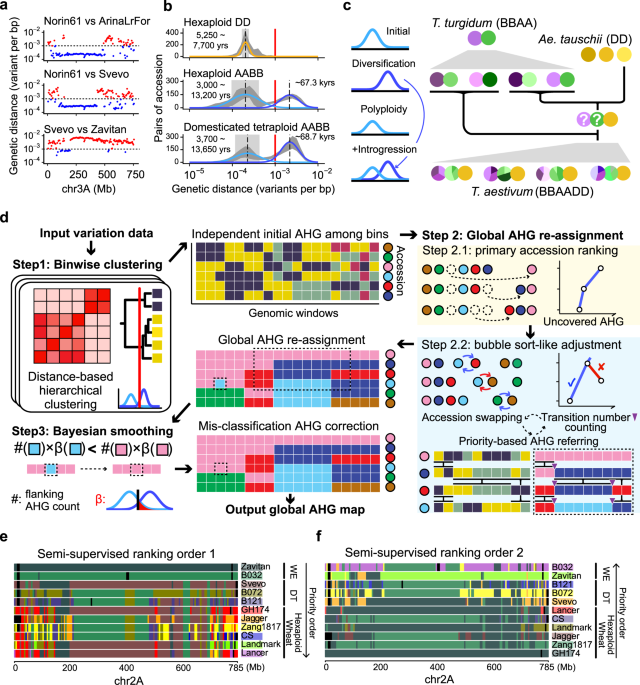
In 2017 November, when exploring the whole genome sequencing data of polyploid wheat, we observed an interesting pattern of the trimodal distribution of the genetic variant densities across the genomic windows. With further evidence, we speculated the lower-density subdistribution represented the genetic diversities cumulated in modern breeding (hundreds of years), the mid-density subdistribution corresponding to the domestication (thousands of years), and the higher-density subdistribution may reflect the different sources of wild emmer wheat lineages, which have been divergent for tens/hundreds of thousands of years.

Model of mixed distributions of genetic variant densities in wheat
By distinguishing the first two components, we developed a novel algorithm called “ggComp”, dissecting wheat germplasm resources at high genomic resolution successfully2, and precisely defining the germplasm-based haplotypes (gHap) with the support of a statistic model. In the recent paper published in Nature Communications3, we distinguished the latter two components and implemented an algorithm "IntroBlocker", which enabled us to trace the gene pool across emmer wheat and common wheat in terms of ancestral haplotype group (AHG). Combining the "IntroBlocker" algorithm and the population-scale sequencing data of wheats, we constructed a pan-ancestry haploblock map presenting the inferred evolutionary trajectories of each genomic window for the polyploid wheat.
The promising approach of polyploid wheat to success
In the recent work3, we showed many new aspects for understanding the success of polyploid wheat as being domesticated to be a widely planted crop.
First, we combined both nucleic-genomic and plastic-genomic evidence and proved that the domesticated tetraploid wheat emerged from the admixture of six main founder lineages of wild emmer wheat dispersed around the Fertile Crescent. The sequence templates (ancestral haplotype) of the major domination loci could be traced to variant wild emmer lineages, indicating the onset of wheat domestication was selecting and mixing the different wild emmer wheat. The admixed genetic diversity contributed to the mosaic genome of the domesticated tetraploid wheat.
Second, the hexaploid common wheat originated from a natural hybridization and polyploidization event between durum wheat and wild goat grass. The common wheat became more successful by spreading along the cultivation of domesticated wheat, leveraging the resourceful gene pools of domesticated emmer wheat through interploidy introgression: tetraploid wheat was speculated to serve as the maternal donor during early spreading, and hexaploid turned to have more chances to be the maternal donor in the late stage for tetraploid-hexaploid hybridization, considering the hexaploid common wheat become dominant. The polyploid nature makes it easy to exchange genetic materials across ploids and hexaploid wheat could utilize the domesticated gene pool of tetraploids directly. "Blue is extracted from the indigo plant, but is bluer than it".
Third, we showed that the “wild-to-crop” introgression and interploidy introgression have played important roles in maintaining genetic diversity during the wheat domestication process.
Fourth, genomic evidences indicated the key domesticated genetic loci were cumulated and fixed gradually throughout this protracted domestication process across tetraploid and hexaploid wheat. It is a ‘smart’ and also ‘economic’ way for wheat to rapidly gather the beneficial genes from different lineages and even from different ploids that have been domesticated in parallel during evolution.

The dispersed emergence and protracted domestication process of polyploid wheat
In summary, as a typical polyploid plant, wheat showed its advantages and special genomic properties in successfully becoming a major crop. Whereas, some special properties might be a two-edged sword for domestication and modern breeding. For example, we identified the long conserved centAHG (Centromeric ancestral haplotype group) zone for each chromosome, which contributed to the diversity of wheat varieties and even sub-species during the domestication and spreading, but would hinder the wheat improvement efficiency in the modern breeding process.
Possible solutions for future challenges in food security
During the past few years, we have focused on the major process of crop evolution, including domestication3, breeding2, and de-domestication4 in the system of wheat, the representative crop important in both evolutionary research and global food security. The knowledge embodied in this series of works has the potential to attribute to the theoretical foundation for accelerating the dissection of germplasm resources and the creation of excellent cultivars. As climate change accelerated, understanding the genomic shaping process during crop evolution will benefit the breeding design and improvement for overcoming future challenges in food security5.

Our recent works on understanding wheat (de-)domestication and breeding
References
1 Dubcovsky, J. & Dvorak, J. Genome Plasticity a Key Factor in the Success of Polyploid Wheat Under Domestication. Science 316, 1862, doi:10.1126/science.1143986 (2007).
2 Yang, Z. et al. ggComp enable dissection of germplasm resources and construction of a multiscale germplasm network in wheat. Plant Physiology 188, 1950-1965, doi:10.1093/plphys/kiac029 (2022).
3 Wang, Z. et al. Dispersed emergence and protracted domestication of polyploid wheat uncovered by mosaic ancestral haploblock inference. Nature Communications 13, 3891, doi:10.1038/s41467-022-31581-0 (2022).
4 Guo, W. et al. Origin and adaptation to high altitude of Tibetan semi-wild wheat. Nature Communications 11, 5085, doi:10.1038/s41467-020-18738-5 (2020).
5 Huang, X., Huang, S., Han, B. & Li, J. The integrated genomics of crop domestication and breeding. Cell, doi:10.1016/j.cell.2022.04.036 (2022).






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in